
Architecture
Overview
BioGUID.org consists of four separate database files, a set of web services, and a website. The database files contain the data indexes themselves, as well as supporting information, and code related to data services that are called internally by the databases themselves as well as by the web services. The web services are the foundation of the BioGUID.org APIs, and serve as the interface between the data services and the website. The website includes a series of pages that include documentation (Overview, APIs, FAQ, and Use Cases), as well as access to data import and export interfaces and the Identifier Domain management system.
Most of the development effort for BioGUID.org focused on the Data Model and associated data services, which is where most of the "heavy lifting" happens. The web services are intended to provide a functional yet relatively light-weight ("thin") web-based interface to the data services, for use by external parties and used internally by the BioGUID.org website. Although never intended as a website per se (the original intention of BioGUID was as a set of data indexes and services; not as a website for direct user interaction), the website has proven useful for storing documentation, and providing very simple user interfaces to some of the core web services. Depending on how BioGUID.org evolves (in terms of user demand, data content, and funding), the website may be improved and expended to include access to more services, be more user-friendly, and function as a full-blown portal to the BioGUID.org system.
The data model is described below, and the web services are described on the API page. The database files and web service and website files are all hosted on a GoDaddy server with an Intel Core i5-2400 processor operating at 3.1Ghz, 32GB of RAM and two 1TB drives, installed with Microsoft Windows Server 2012 Standard. The database platform is Microsoft SQL Server 2014 (v12) Standard Edition, and the website and services are operating on Apache Tomcat 7.0. All source Code is or soon will be deposited on the BioGUID GitHub site.
Databases
At the heart of BioGUID.org are four separate database files. These were created and are currently maintained on the Microsoft SQL Server DBMS platform (though they could easily be ported to a different relational database platform, such as MySQL). All code (Stored Procedures, Functions and Views) are written in TSQL, and as much as possible they avoid the use of any proprietary features proprietary to Microsoft's implementation. The four database files in BioGUID.org include:
- BioGUID [Core Data Indexes]
- BioGUIDDataServices [Views, Stored Procedures, Functions and support data tables]
- BioGUIDLocalLog [Local performance and usage statistics monitoring]
- BioGUID_IMPORT [Database used for bulk importing of data]
BioGUID
FileName: BioGUID.mdf Size: 550GB Tables:
- DereferenceService – Each row represents a Dereference Service (service used to dereference or "resolve" identifiers over the internet).
- EditLog – Stores information on all data changes to managed tables, included which field of which record of which table changed from what old value to what new value, when, by whom, and in some cases, why.
- Enumeration – Table for storing all controlled vocabularies within BioGUID.org.
- IdentifiedObject – Each row represents a conceptual object, to which one or more identifiers are assigned.
- Identifier – Each row represents a single identifier, issued by an Identifier Domain, and applying to a conceptual object listed in the IdentifiedObject table.
- IdentifierDomain – Each row represents an Identifier Domain, from which identifiers are issued.
- IdentifierDomainDereferenceService – Join table establishing the many-to-many relationship between Identifier Domains and Dereference Services.
- PK – A universal table managing core record information for each record in the managed tables (i.e., all tables except Identifier, IdentifiedObject, and ResourceRelationship).
- ResourceRelationship – A table that establishes non-congruent relationships among conceptual objects represented in the IdentifiedObject table.
- SchemaItem – A table used to store information about each table and field in the BioGUID database.
- view_FieldDetails – Provides schema information on fields. Used for internal processes.
- view_FKSubtypes – Lists tables as Subtypes (1:1). Used for internal processes.
- view_PKFields – Determines the primary key field(s) of tables. Used for internal processes.
- sp_CompareValues – Used for comparing field values at a binary level (i.e., independent of collation settings).
- sp_CreateCache – Used for establish the Cached values (used by Full text Indexing) for IdentifiedObjects.
- sp_FTIStatus – Used to determine the current status of any Full-text indexing processes.
- sp_GetCurrentUser – Determines who the current user is. Not currently used, but will be when authentication is added to BioGUID.org.
- sp_PerformanceLog – Used for analyzing performance characteristics of various stored procedures.
- sp_ProcessEdit – Processes edits from triggers in database.
- sp_SynchronizeSchemaItemDescription – Synchronizes the Description field of the SchemaItem table with the stored description metadata for the corresponding table or field in SQL.
- sp_UpdateCache – Used for refreshing the Cached values (used by Full text Indexing) for IdentifiedObjects.
- CleanSearchTerm – Prepares search term for use on searches (e.g., to remove excess whitespace and avoid injection SQL attacks).
- FormatFullIdentifiers – Formats array of identifiers for object Cache field.
- GetPKFieldName – Used internally to establish the field name for the primary key field of a table.
- GetRecordCount – Used to get the record count quickly on large tables.
- GetSchemaItemID – Used to convert the name of a SchemaItem (Table name or field name) into its corresponding internal ID value.
- EditLogPackage – User-defined Table Type used for transferring record edits to the EditLog manager.
- PKIDListType – User-defined Table Type used for improving performance on indentifier searches.
| Figure 1. |
|---|
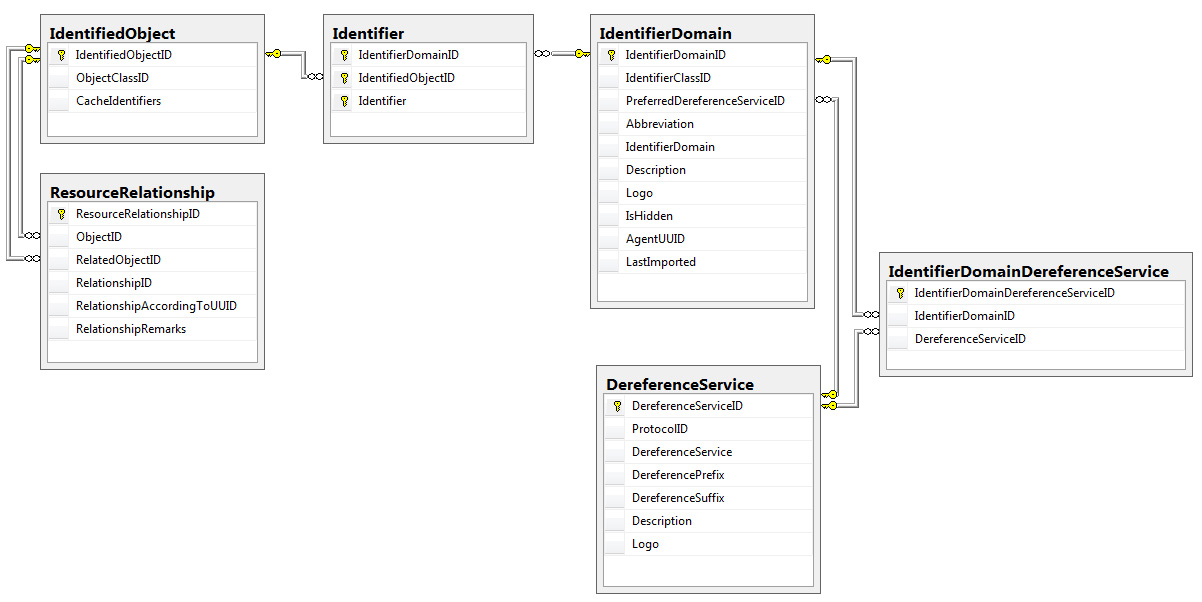 |
| The relationships among core data tables in the BioGUID database. |
| Figure 2. |
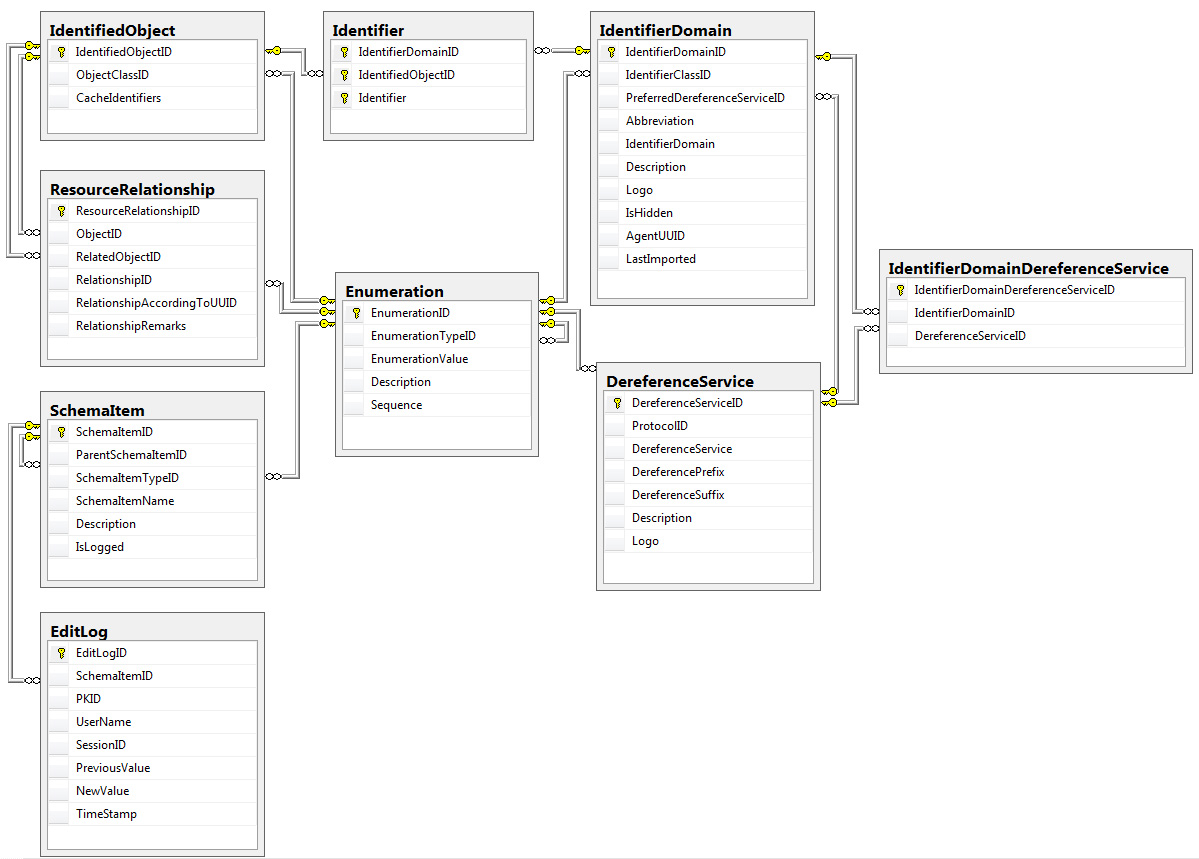 |
| The relationships among core data tables and supporting data tables in the BioGUID database. |
BioGUIDDataServices
FileName: BioGUIDDataServices.mdf Size: 0.5GB Tables:
- FAQ – Table that stores items displayed in the website FAQ page.
- FilterTerm – Table that stores certain search terms that are commonly entered by robots fishing for security holes.
- NewsItem – Table that stores items displayed in the website home page news feed.
- SearchIndex – Table that temporarily stores results of searches, used for enhancing performance.
- Temp_MultipleIdentifiers – Table that temporarily stores results from multiple identifier analysis on large datasets.
- view_BatchImportStatus –
- view_GetIdentifierDomainDereferenceService –
- view_MultipleIdentifiers –
- view_SearchDereferenceService –
- view_SearchFAQ –
- view_SearchIdentifier –
- view_SearchIdentifierDomain –
- view_SearchNewsItem –
- sp_AdjustCreatedUsername –
- sp_CompareValues –
- sp_ConvertCRTABtoSpace –
- sp_ConvertEmptyToNull –
- sp_DeleteRecord –
- sp_EditValue –
- sp_EXPORTIdentifier –
- sp_EXPORTWithinDomainMultipleIdentifiers –
- sp_GetBatchImportStatus –
- sp_GetEXPORTStats –
- sp_GetIdentifierDomainDereferenceService –
- sp_GetSearchDetails –
- sp_GetStatistics –
- sp_IMPORTBatchTransfer –
- sp_IMPORTDuplicateObjects –
- sp_IMPORTIdentifierProcess –
- sp_IMPORTIdentifierTransfer –
- sp_InsertDereferenceService –
- sp_InsertEnumeration –
- sp_InsertIdentifierDomain –
- sp_InsertIdentifierDomainDereferenceService –
- sp_InsertNewsItem –
- sp_InsertSchemaItem –
- sp_LookupDereferenceService –
- sp_LookupIdentifierDomain –
- sp_MergeObjects –
- sp_MergeObjectsSimple –
- sp_NewPKID –
- sp_PerformanceLog –
- sp_PerformanceSummary –
- sp_SearchDereferenceService –
- sp_SearchEnumeration –
- sp_SearchFAQ –
- sp_SearchIdentifier –
- sp_SearchIdentifierDomain –
- sp_SearchNewsItem –
- FullEnumerationChildList –
- FullEnumerationParentList –
- CanonicalIdentifier –
- FormatAlternateDereferenceServices –
- GetDereferenceServiceID –
- GetEnumerationID –
- GetIdentifierDomainID –
- GetItemID –
- GetParameterValue –
- GetPKID –
- GetSchemaItemID –
- GetUUID –
- IsUUID –
- NormalizeDOI –
- NormalizeISSN –
- NormalizeUUID –
- ParseSearchTerm –
- RemoveWhitespace –
- StripDiacritics –
- StripHTML –
- DupeIOType –
- IDResponseType –
- PKIDListType –
BioGUIDLocalLog
FileName: BioGUIDLocalLog.mdf Size: 0.8GB Tables:
- PackageLog – Table that stores data packages transmitted to the EditLog manager system.
- PerformanceLog – Table that stores performance characteristics of stored procedures.
- SearchLog – Table that stores a log of searches made on BioGUID.org.
- SearchResult – Table that stores the result-set from searchers made on BioGUID.org.
BioGUID_IMPORT
FileName: BioGUID_IMPORT.mdf Size: 19GB Tables:
- IMPORT – Table that stores post-processed records from bulk import procedures.
- IMPORT_Temp – Table that stores pre-processed records from bulk import procedures.
 |
All content within the BioGUID.org site is available under the Creative Commons Zero license (Public Domain). |